
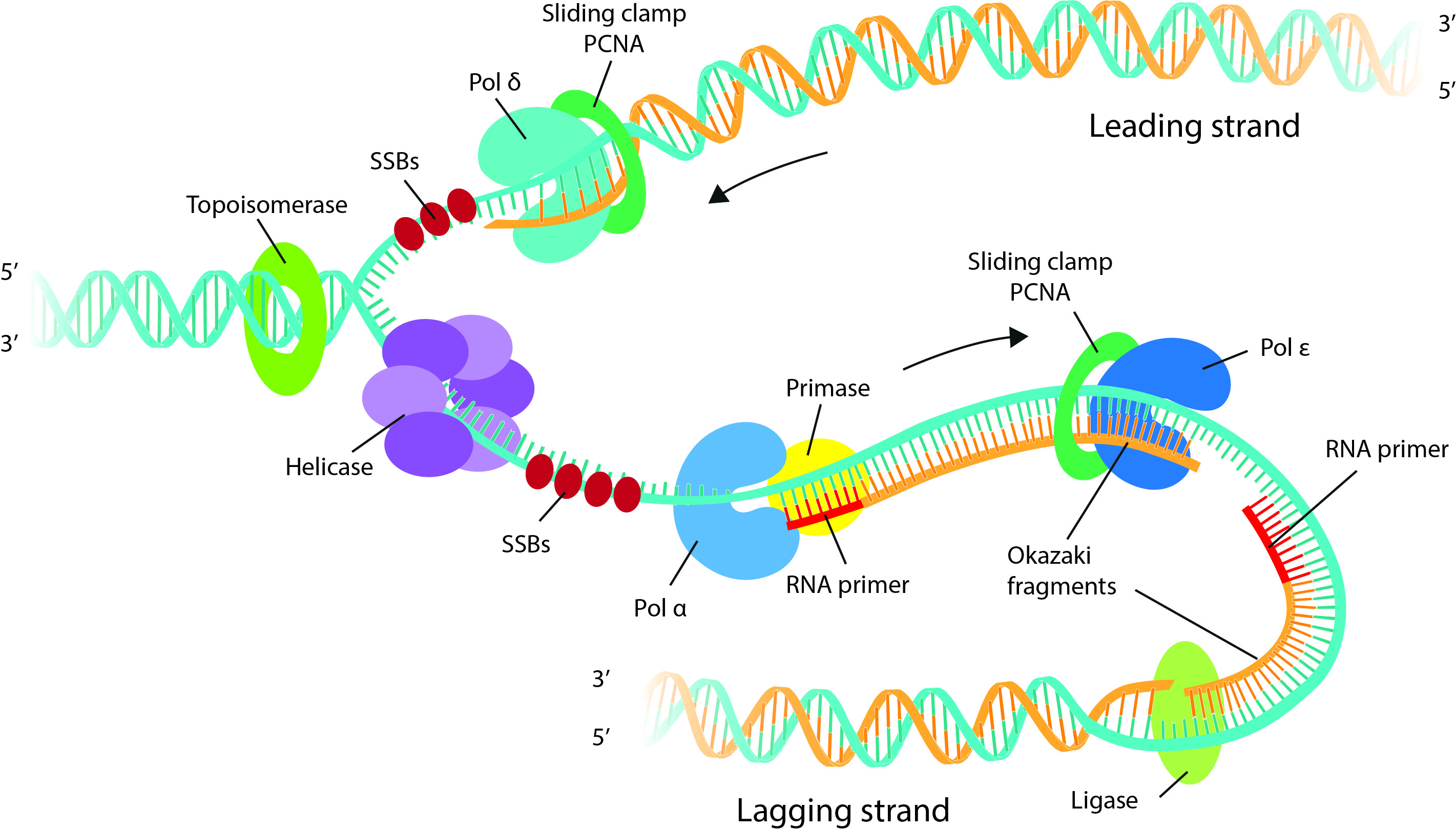
Because each cell has multiple copies of the TCR or BCR transcript, RNA-seq approaches are more sensitive. Using genomic DNA as a template can make it easier to determine the relative abundance of clonotypes, as each cell only has a single template. What factors should I consider when choosing an immune repertoire sequencing approach? DNA vs. Along with this added technology in the researcher's toolkit comes several key decision points on how to proceed with your studies. Although these methods have yielded valuable insights, the continued development of next-generation sequencing (NGS) has dramatically expanded the scale of repertoire profiling research. Traditionally, immune repertoire profiling has relied on cloning and Sanger sequencing or functional methods for identifying antigen-specificity (e.g., tetramer assays/stains). However, it is this very diversity of receptor sequences that can make it difficult to analyze them, and a myriad of research applications-from examining the mechanisms behind autoimmune diseases and cancers to developing models for immunotherapy-rely on being successful in this first step.
Ultimately, this process yields cell populations with sufficient TCR and BCR diversity to recognize any peptide imaginable. To this end, the adaptive immune system has evolved a system for somatic diversification, commonly referred to as V(D)J recombination. Receptor-antigen interactions are specific, meaning a tremendous amount of diversity in TCRs and BCRs is required to recognize the wide assortment of antigens one might encounter. Adaptive immunity relies on the ability of the body to recognize and respond to a nearly endless number of molecules, a complex feat achieved through T-cell and B-cell receptors (TCRs and BCRs, respectively).


 0 kommentar(er)
0 kommentar(er)
